Introduction
In recent years it has begun to become apparent that the methods currently used in designing, evaluating and implementing public health policy do not work (Bruch & Atwell 2015). This is especially the case in epidemiology, for example, epidemiological forecasting models for fatal diseases such as AIDS have been known to fail because individuals adapt their behaviour to the epidemic (Epstein et al. 2008). One of the reasons for this type of failure is the reliance on equation-based models to model the spread of an infectious disease in a population. Equation based models are relatively simple to implement and have been shown to produce results that generally mimic that of an outbreak scenario (Skvortsov et al. 2007), however, the models assume that the population being modelled is homogeneous (Friás-Martínez et al. 2011) which is a serious limitation. In order to include multiple populations, an equation-based model requires a large number of complex equations and each subpopulation or category in the model will have a separate set of parameters. This will make a model very complicated and difficult to understand and solve.
Agent-based simulation models are an alternative to equation-based models and can model much richer scenarios than the latter. An agent-based simulation model allows agents to interact with other agents and the environment based on a set of rules (Bruch & Atwell 2015) and agent-based models can be an essential part of an epidemiological study when it is not feasible to run an experiment (Hernán 2014). For example, agent-based simulation models can show what happens in cases where there is failure in uptake of measures of disease prevention (Homer & Hirsch 2006).
With an agent-based model each individual agent can be given different attributes and make different decisions. This allows the model to capture interactions and behaviour at the individual level (Bobashev et al. 2007). In the case of epidemic models agent-based models allow for high fidelity modelling on a global, national and community scale (Skvortsov et al. 2007). Individual mobility and social networks are essential parts of the spread of an infectious disease and agent-based models are able to capture these interactions when an equation-based model fails to (Friás-Martínez et al. 2011).
One advantage of using agent-based models over approaches such as equation based models is that agent-based models allow for more flexibility and a large amount of freedom in terms of the simulation design. This level of flexibility and freedom comes with a cost: many models in the literature propose their own agent interaction structure (Richiardi et al. 2006). Combined with the application to many fields this leads to a wide range of agent-based models and descriptions (Friás-Martínez et al. 2011). This leads to difficulty in finding a general definition for agent-based models. To combat the issue of definition, for the purpose of this review we define an agent-based model as a computer simulation that involves agents in an environment with a time and spatial component. Agents must interact with each other and with the environment. One important feature of an agent-based model is that the agents differ. The difference can be in assigned attributes such as age, gender, or health status; or the difference can be in how the agents act. The actions of agents are governed by a behaviour control program. At each time step an agent decides what it will do: the actions can be as simple as defining which direction an agent will move in based on some simulated perception or the actions can be more complicated such as searching for agents with certain characteristics within a given radius and interacting with them (Mac Namee & Cunningham 2003).
Because of their ability to model individual behaviours and interactions, agent-based models are well suited to epidemiological simulations, especially those involving infectious diseases. In our review we focus on infectious disease epidemiology. The majority of diseases that are handled in the simulations are acute diseases, however there are some infectious diseases in our review that lead to chronic conditions. As we are only focusing on the initial infection and spread of the disease, we do not consider models simulating chronic disease.
Because there is no set methodology for agent-based models there is a wide variety of epidemiological agent-based models with different levels of detail, results and uses. This makes it difficult not only to understand and compare different models and also makes it difficult for researchers to know where to start when first creating their own simulations. A taxonomy of agent-based infectious disease models could help in both of these situations. To our knowledge, at the present time no work has attempted to create a taxonomy for the existing epidemiological agent-based models. This literature review will analyse the current body of work on epidemiological agent-based models with the goal of classifying the existing simulations and to create a taxonomy for agent-based epidemiological models.
This paper will proceed as follows. Section 2 will review the existing literature on agent-based epidemiological models and will discuss the important components that need to be considered when creating an agent-based epidemiological model for infectious diseases. In Section 3 we will use the knowledge of agent-based epidemiological models we gained from performing the literature review to create a taxonomy for agent-based epidemiological models for infectious diseases. Section 4 will show how the taxonomy can be used to classify models and how it can be used to help researchers in creating their own models.
A Review of Epidemiological Agent-based Models
Agent-based models are an important tool in studying the dynamics of infectious diseases. In many cases it is impossible to run an experiment to see how a disease will affect a population in the real world so an agent-based model can be used instead. Simulations are already being used to help decide on policy in the models by Barrett et al. (2008), Aleman et al. (2011) and Lee et al. (2008). Other models are being used to understand past outbreaks to be better prepared in the future such as Merler et al. (2015)’s model of the Ebola outbreak in Liberia and Friás-Martínez et al. (Friás-Martínez et al. 2011)’s model of the H1N1 outbreak in Mexico City.
As new infectious diseases emerge, agent-based models can be used as an aid to help understand how a population can be affected and how we should react to an outbreak. To do this it is necessary to have a strong understanding of all possible factors in disease spread. Much of the research being done now with agent-based models helps us to get to that point. For example, Epstein et al. (Epstein et al. 2008)’s work on fear leading to agents fleeing the area of an outbreak and spreading the disease further could play an important role in future simulations of emerging diseases.
There are four main components of an epidemiological agent-based model: disease, society, transportation, and the environment. When creating an agent-based model for infectious disease epidemiology one must consider how they will model each of the four components. In modelling the disease it needs to be determined how the infectious disease is transmitted between agents and how the disease progresses in an infected agent. Modelling society involves simulating the population while modelling transportation determines how the agents will move through the environment. Modelling the environment involves creating the space in which the agents will interact. Although we separate them for the purpose of understanding the agent-based epidemiological model, the components are intertwined. For example, modelling the disease will determine if a susceptible agent who comes into contact with an infected agent becomes infected, however the contact between agents is determined by the how transportation is modelled. In reviewing the literature we focused on how different models treat these four main components, and how they papers present model validation.
We find common themes in the literature on how the four main components are dealt with. For example, creating a more general model that can easily be adapted to multiple diseases versus creating a more specific model for a given disease. As detailed more in the sections below, different methods have both advantages and disadvantages. While specific models are often more robust and based on more extensive data they can take more time to run and are less adaptable. General models while adaptable are sometimes to simple and cannot be related to a real world scenario. It is the right combinations of specific and general components that makes a successful model. The combinations along with their advantages and disadvantages are discussed in the taxonomy section. For example, a general disease and general society model are simple to create and run, but do not produce results that can be used for anything besides simple disease dynamics research.
Validation is an important part in designing any model. However, it is not always clear how to validate an agent-based model. Although some authors attempt to detail their validation process, many papers fail to mention validation at all or only briefly mention that it was done. The methods or lack of methods used to validate the different models are described in the validation section below in more detail.
Modelling Disease
The agent-based modelling literature tends to treat infectious diseases in one of two different ways. Research is either done to create a general model where the disease parameters can be changed to show how various diseases will spread through different populations or the research focuses on modelling a specific disease and often a specific outbreak of that disease. A general disease model should be adaptable to multiple diseases of the same form of transmission, typically airborne transmission. These general disease models make sense in a scenario where the modellers want to create a tool to study future potential outbreaks. This way the model can be adjusted to different disease dynamics based on what disease is to be studied without creating a new model each time. For example, Barrett et al.’s (2008) Episimdemics was created for the US without a specific disease so that it could be adjusted for different possible outbreaks. It has been used to help determine policy in the face of a pandemic in the US.
A specific disease model allows for a model to better capture specific disease dynamics. While general disease models typically stick to airborne transmission, specific models can take into account other transmission methods such as water borne infections. Specific disease models can also take into account factors that might influence the spread of a given disease such as including infection during funerals in the model of Ebola spread in Liberia by Merler et al. (2015).
Regardless of the specific or general nature of the model, the disease model will have many of the same components. The breakdown of the components can be seen in Figure 1. Disease models for agent-based models are broken up into two parts: between host transmission and within host progression. Between host transmission occurs when a susceptible agent comes into contact with an infective agent, and the between host transmission component of a disease model simulates how a disease is transferred when this occurs. The within host progression component of a disease model simulates how, when an agent becomes infected, they move between the different states of the infection (for example exposed, infective, and recovered). Both parts of the disease model are important in accurately simulating how a disease will spread.
The transmission dynamics are a key factor in how the disease spreads between individuals. Disease can be spread through human-to-human contact, to humans from food or drinking water, or between hosts of different species, for example mosquitos to humans. When an agent comes into contact with an infected agent, infected food or drinking water or an infected species, a probability distribution is used to determine transmission. The transmission can be affected by a number of factors outlined in Figure 1: transmission dynamics, society, transportation and environment, and behaviours. A number of agent-based models dealing with specific diseases contain different transmission dynamics based on the disease being modelled. Some of these models are for diseases such as cholera or malaria that are not spread through human contact but through contaminated food and drinking water or through insect bites. In the model by Crooks & Hailegiorgis (2014) for the spread of cholera in a refugee camp agents excrete a certain amount of the cholera bacteria based on the stage of infection that they are in. Water contamination is determined through a hydrology model where the flow of rain water affects the total amount of pollutant and if an agent drinks contaminated water they have a certain probability of becoming infected. Malaria is spread by mosquitoes thus any simulation for malaria would have to take into account not only human movements but also mosquito movements (Linard et al. 2009). In addition, models can alter different transmission dynamics by agents for example by making some agents super spreaders, individuals who are more infectious than other individuals (Duan et al. 2013).
The parameters of the society being modelled have a major effect on how a disease will spread between hosts. A densely populated area will result in more contacts between agents, and thus a greater likelihood of infection (Perez & Dragicevic 2009). The social networks of agents also influence the disease spread. In their model of a disease spreading through a small Australian town, Skvortsov et al. (2007) found that the majority of infections in their model occurred at the schools. This was because every agent at a school was in contact with every other agent at the school. The large social networks of agents in the model led to a higher infection rate.
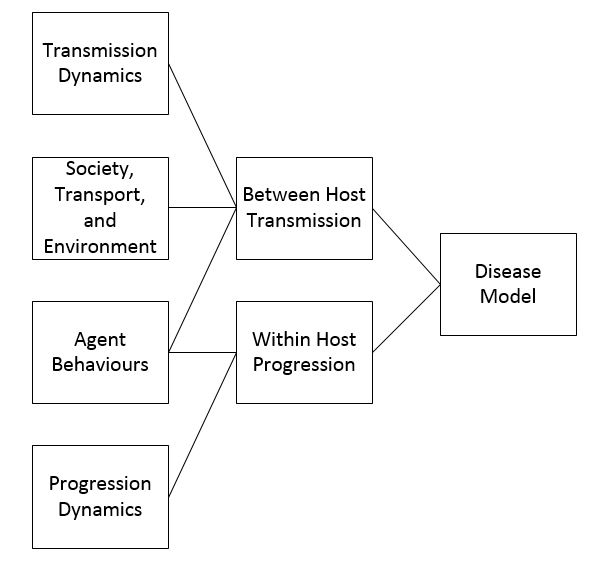
Agents’ behaviours can also have an effect on the between host progression of a disease. For example, if the agents respond to an outbreak or possible outbreak by fleeing they may be spreading the disease at a greater rate than if they stayed home in isolation (Epstein et al. 2008). Alternatively, if agents choose to isolate themselves once infected, they reduce the number of contacts they make and thus reduce the number of agents the disease spreads to (Dunham 2005). Diffusion of information about a disease can lead to agent’s taking part in preventative behaviours such as getting vaccinated or taking medicine, such as flu prophylaxis, that will reduce the chance of infection (Mao 2014). When modelling the Ebola epidemic in Liberia, Merler et al. included change in agents behaviours based on information about the disease. In the real epidemic as people learned that Ebola spread at funerals and to health care workers and other non-Ebola patients in hospitals the number of safe funerals increased and health centres that only treated Ebola patients opened thus reducing transmission of the virus. This was reflected in the model with the number of hospital beds changing as time went on and the probability of becoming infected at funerals decreasing (Merler et al. 2015).
Within host progression does not have as many outside influences as between host transmission. The make-up of the society has no effect on how an agent moves from exposed to infected or infected to recovered. Similarly behaviours of other agents have no effect on within host progression. While an infected agent deciding to stay home from school or work might reduce the chances of other agents becoming infected, other agents’ actions will have no effect on the progression of a disease within an agent (Mao 2014). On a basic level all of the within host progression models are similar. A disease will move between states based on a probability distribution. Many of the agent-based models use a form of the SIR model to simulate disease progression (Dunham 2005; Friás-Martínez et al. 2011; Merler et al. 2015; Perez & Dragicevic 2009; Rakowski et al. 2010; Crooks & Hailegiorgis 2014). The SIR model categorizes individuals into susceptible, infected or recovered states and looks at movement of individuals between these. Variations of the SIR model can include additional stages such as exposed (Keeling & Rohani 2008). Although the SIR form of the disease model can be used for a specific disease it is often used when a simulation is created for a general disease. A few models take more complicated disease dynamics into account moving away from the basic SIR type model. In modelling tuberculosis (TB), Tian et al. (2013) include states particular to TB including high and low risk latently infected agents, latently infected with previous treatment agents, undiagnosed infectious and non-infectious agents, active TB agents, and active undiagnosed infectious and non-infectious TB with previous treatment agents.
Although other agents do not have an effect on within host progression, the infected agent’s behaviours can have an effect on the progression. Preventative behaviours can reduce the chance of an agent moving between susceptible and infected or increase the chance of an agent moving between infected and recovered (Mao 2014). In some models having been vaccinated can reduce the chance that an agent moves from exposed or latently infected in the case of tuberculosis (Tian et al. 2013).
The factors affecting the transmission or progression can vary between the models but typically fall into the categories of progression dynamics, behaviours and society factors. However, the general disease models will have simple transmission and progression models while specific models tend to have more complicated transmission and progression models that reflect the given disease. The more factors added into the model the more realistic it will be. This, however, comes at a price and increased model complication leads to increased computational resource requirements.
Modelling Society
In the spread of an infectious disease one of the main components that can have an effect on the course of the outbreak is the structure of the society. The number of people or agents, the household structure, number of students in each school, number of schools and workplaces are all things that need to be considered when simulating a society. It must be determined if the model will simulate an existing society or if it will be more general. We consider any simulation that uses real data to model a society a specific society model and any model that generates a society without the use of real data a general society model. General society models can be made by randomly placing agents in an environment. For example, the model by Dunham (2005), was created by generating 50 genderless and ageless agents and having them commute back and forth from their home locations to their work locations. Similarly, Perez & Dragicevic (2009) created a society by randomly assigning genderless and ageless agents to a residential area and then randomly dividing that population into workers and students. The advantage of creating a general society model is that it does not require the large amounts of data necessary to simulate a real society. Because the data is not needed it will take less time for a modeller to begin the process of creating the simulation and the initialization of the simulation will need less computer power and time.
In order to create a simulated society model based on a real society, simulation designers typically take census data from the population they are planning on recreating or from a similar population. For example, Skvortsov et al. (2007) used census data to determine the age/sex breakdown of the actual population of an Australian town and had the model build families based on average family size obtained from the census data. To model the spread of influenza through Poland, Rakowski et al. (2010) use census data to assign individuals to a family based on age and relationships: a child will only be assigned to a house if an adult is already living there and the probability that two adults will live in the same house depends on the attraction which is determined by the difference in age between them. The scale of a specific society simulation can range from a small town (Skvortsov et al. 2007), to a community (Lee et al. 2008), to a region (Aleman et al. 2011) or to a country (Ajelli et al. 2010). As it is important to capture the social networks of an individual to determine the path of a disease spreading through a society, some models can differentiate between close contacts (other agents at home or work) and occasional contacts (agents in service places such as shops) (Crooks & Hailegiorgis 2014). The social networks of agents in the society can also be broken down into weekday and weekend networks as it is more likely that an agent will interact with co-workers during the week and with friends during the weekend (Friás-Martínez et al. 2011).
Specific societies have a more obvious interpretation and use: their results can be applied to a given population to help make decisions about future outbreaks or learn from past outbreaks. However, in order to create such a simulation data is needed and the more realistic a simulation is the more detailed data that is required. General society simulations may not require any data at all and thus can be easier to create in situations where data is scarce or hard to access.
The scale of the model must also be considered: for a simulation of a specific society the scale (country, region, city etc.) will be determined by the society being simulated. However, for a general model it is necessary to determine how many agents will be used in the simulation. The scale chosen for the society can also have an effect: the larger the scale the more computing time the simulation will take to run. However, small scale societies, particularly small scale general society simulations, may not have as much realistic interpretability as it would be difficult to find a real world application for such a model. The way that society is simulated will influence the rest of the model including how transportation is simulated and how the results of the model will be interpreted and used.
Modelling Transportation
The majority of agent-based epidemiological simulation models contain some form of movement or transportation of agents through the model environment, and choices must be made about how to simulate this. The majority of simulations drive agent movements based on the society model and the agent behaviour rules. Typically an agent will simply move from the house to which they are assigned to their workplace every day. Some more sophisticated models, however, also include destinations such as markets, shopping malls, pubs, friends’ homes, health centres and religious centres (Crooks & Hailegiorgis 2014; Mao 2014; Perez & Dragicevic 2009; Simoes 2006) and simulate agents movements between these locations following a weekly schedule.
The transport model in a simulation governs the way in which agents move between different locations. Simulations can use a very simple transportation model where agents simply move between locations in a straight line at a constant speed (Dunham 2005). More realistic transport models use geographic data containing information about transport infrastructure to plan routes between destinations following footpaths and roads. Some models require the agents to select the shortest route (CITE) while others allow less optimal travel (Crooks & Hailegiorgis 2014; Perez & Dragicevic 2009). It is possible to have more specific data to model movements such as cell phone data where an individual’s real movements can be tracked based on where a phone call or other telephone service is used (Friás-Martínez et al. 2011). However, this kind of data is not easily accessible to all researchers. Some of the most sophisticated transport models include public transportation, as public transportation can be a crowded location where diseases are transmitted (Rakowski et al. 2010; Aleman et al. 2011).
Movement can be affected by the agent’s choices and behaviour. For example, if an agent is infected a model can allow the agent to decide if they are going to take a sick day (Dunham 2005). A model by Crooks & Hailegiorgis (2014) went further allowing agents to set goals based on an agent’s attributes and needs that determine movement. Travelling longer distances can also be considered. A model for the spread of mumps in Portugal (Simoes 2006) not only considers neighbourhood and intra-region travel, but also has a component for inter-region travel. In modelling influenza epidemics in Poland, Rakowski et al. (Rakowski et al. 2010) assign a certain number of agents at each time step to traveller status. These agents then choose their end points, transfer cities, and co-travellers. Co-travellers and the number of co-travellers are chosen randomly to simulate both public and private transportation. The movements of agents can have a great effect on the outcome of a simulation. Movements will determine who an agent contacts and thus affect how a disease will spread. Some of the advantages of including transport in the model is the ability to capture the location of infections. This could help identify potential ’hotbeds’ of infection such as schools. It also allows for more realistic interactions outside of an agent’s family or friends network.
There are some infectious disease models that do not include transportation of any kind. These models rely on contact networks to determine the spread of the disease. Agents who are in networks with other agents have a probability of coming into contact and spreading the disease. Tian et al. (2013) use such a model for their TB analysis and Olsen & Jepsen (2010) similarly create a model without transportation to model the spread of HPV. If the disease dynamics are not as reliant on agents’ day to day movements then not including transportation in the model will lead to a faster run time for the model.
Modelling the Environment
The environment is an essential part of the agent-based model as it is where the agents move and interact. However, the level of complexity of the environment can vary based on the needs of the model and the transmission dynamics of the disease. An environment model can be as simple as a spatial grid upon which agents are placed as in Duan et al. (2013) and Dunham (2005). Such as environment can be seen in Figure 2. Simple environment models are easy to set up and run, however, they give little more information on the contact patterns of agents then you would get from a equation based model while models with added environment are more capable of capturing heterogeneous mixing. Slightly more complicated environments can include buildings for workplaces and schools, roads, and residential areas. An example of this type of environment from Mao (2014) is shown in Figure 3. Lee et al. (2008) creates an environment that includes schools and residential areas. These models have an advantage over the grid environment model as realistic movement patterns can be simulated and high infection areas can be determined. Lee et al. use their model to determine when school closing policies should go into effect and how long the closings should last. Such analysis could not be done with a simple grid environment: the inclusion of the schools and workplaces allows the modellers to accurately capture where agents become infected. Along with the buildings and roads, many models also use other GIS data including elevation data to create their environment such as the models by Ajelli et al. (2010), Barrett et al. (2008) Crooks & Hailegiorgis (2014) and Simoes (2006). This allows for an accurate representation of the town, city, country etc. that is being modelled. Figure 4 shows the environment in Crooks and Hailegiorgis’s model with elevation data.

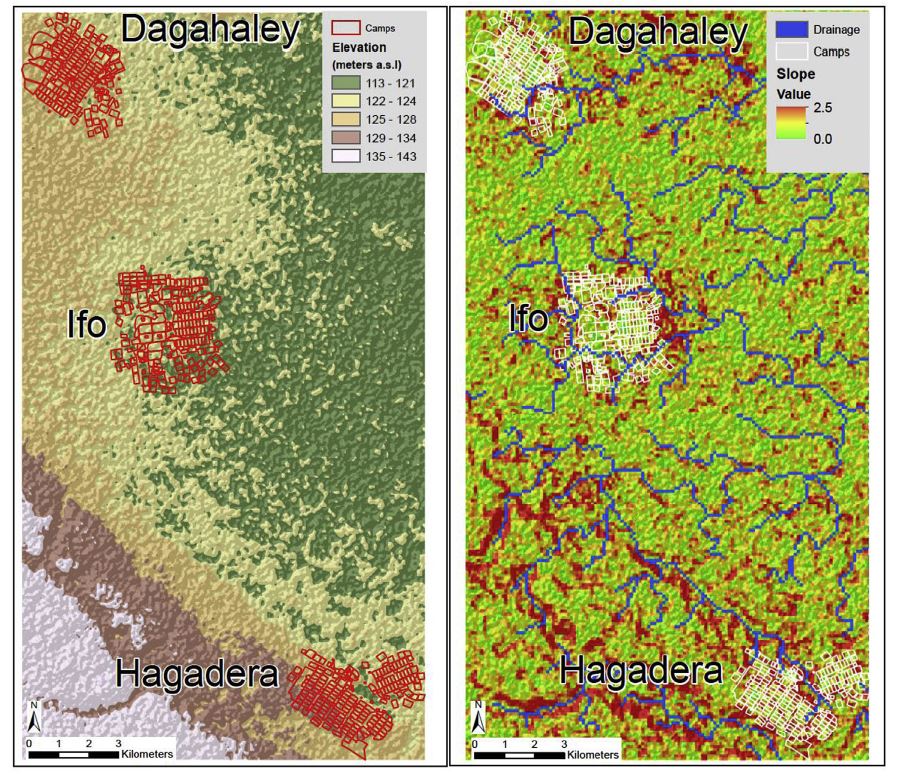
However, some models require a more detailed environment beyond roads and buildings as the environment can have an impact on disease transmission. Sophisticated environment models can also include factors such as temperature or precipitation or other populations that help to spread a disease can be included in the model. Linard et al. (2009) not only include mosquitoes in their malaria model but also temperature, water levels and vegetation levels. In order to model the spread of cholera through a refugee camp, Crooks & Hailegiorgis (2014) include a hydrology model as cholera is a disease spread through the consumption of infected water. Adding the additional environmental factors is essential to model some diseases, such as cholera and malaria, but the transmission dynamics of other infectious diseases can also be influenced by environmental factors. For example, influenza outbreaks most often occur in the winter months. Including environmental factors in an agent based model may capture factors in disease transmission that may have otherwise been ignored. However, the more factors that are included in the model the more complicated it becomes. As agent-based models tend to be computationally intensive additional factors can lead to difficulties in running the model.
Model Validation
One of the most important issues for agent-based modelling is validation. If a model is not validated, any surprising results cannot be completely trusted. There is currently no exact definition or methodology to test the validity of an agent-based model (Richiardi et al. 2006). For epidemiological models, it is possible to simulate an infectious disease outbreak that has occurred in the past. In these cases validation can be possible through comparing the simulated outbreak with the real outbreak (Olsen & Jepsen 2010; Merler et al. 2015; Crooks & Hailegiorgis 2014; Perez & Dragicevic 2009). This gives confidence that the model correctly simulates the dynamics of the disease and the society, allowing the researcher to make the assumption that the model will simulate future outbreaks correctly providing insight into the disease behaviour. For a common infectious disease such as influenza, data sources such as Google flu trend statistics can be used to supplement lab-confirmed reported cases in validation as the Google data will pick up some cases that are not reported (Mao 2014). It is possible to use other statistics besides prevalence to validate an agent-based model. For example, if real movement data is available a comparison can be made between the real movements of individuals and the movements of agents (Friás-Martínez et al. 2011).
However, if the model is not simulating a past outbreak or epidemic (which is often the case with a general disease model) there are ethical, logical, and practical constraints to getting data for validation: it is not feasible to run an experiment to determine how an infectious disease will spread through a population (Hernán 2014). One alternative to using real data for validation is cross validation: the output of an agent-based model can be compared to the output of another widely used model such as an equation-based SIR model. The number of susceptible, infected and recovered individuals over the simulated period can be compared between the two models. Although it is likely that the numbers will not match exactly due to differences in model assumptions, if the infection curves, representing the number of susceptible, infected and recovered individuals at each time step, follow a similar trajectory it is likely that there is some validity in the agent-based model. If a simple agent-based model is validated with equation-based models it is possible for the researcher to add additional factors into the agent-based model. The results of the expanded model can be analysed knowing that the basic disease dynamics of the model were validated (Skvortsov et al. 2007; Rakowski et al. 2010). One other alternatives to validation is determining adequacy (Apolloni et al. 2009). Adequacy is the idea that the appropriate and informed decisions are made when creating the model. When considering adequacy it is important to question if any new input in the model decreases uncertainty and if the input significantly changes the model (Xia et al. 2013).
Although validation is an important step in creating any model, authors do not always include the validation process in their papers. For example, Barrett et al. (2008) mention that the model has been validated but do not describe the validation process. Some papers such as Dunham (2005) do not refer to validation at all. While other papers only briefly mention the comparison to real data in the results or discussion section of the article such as in Crooks & Hailegiorgis (2014) and Perez & Dragicevic (2009).
Designing a Taxonomy
Based on our analysis of the literature we found that for each component there are different categories of agent-based epidemiological infectious disease models. For example, simulations could have a specific or general disease model. These categories can fit together in different ways: such as a general society component being paired with a specific disease component. To better understand how the models fit together we used the descriptions for different levels of the modelling diseases, society, transportation and environment from our review to create taxonomy. The taxonomy can be used to classify the 20 models in our review and can also be used to help classify additional models. Although validation is an important step in the agent-based modelling process we do not include it in our taxonomy because we focus on the components of the simulation instead.
Taxonomy
In order to understand a simulation and its potential uses it is important to note how the components are combined. A taxonomy of epidemiological simulation models based on the level of specificity of the disease, society, transportation and environment model can be created to aid in the classification of agent-based models for human infectious diseases. A taxonomy can help researchers understand where to place their model in the body of existing work and can help them choose what level of complexity they need in their model.
Figure 2 illustrates the taxonomy we propose. The models are then broken down into two categories: general and specific disease models. Those two categories are then broken down to general or specific society simulations, if the model includes a transportation model, and the environmental factors in the model. The grey branches and boxes of the taxonomy that are outline in grey are those combinations of component types that we did not find in our literature review and based on an analysis do not think would be feasible combinations. For example, we do not feel it would makes sense to have an environment made up of maps or maps plus other factors if the model has a general society model. This is because the maps that would be used would be based on the society that is being modelled and in this case no specific society would be modelled. The following sections will describe the different components in the taxonomy.
Disease Model
The taxonomy as seen in Figure 5 breaks the disease model component of a simulation down into specific versus general disease models. General disease models are those that use basic SIR disease dynamics in the model with parameters that can be adapted for various airborne diseases. Simulations with a general disease model prove useful for planning and creation of policy as only one simulation needs to be created to analyse the effects of different outbreaks. The general disease model also allows researchers to save time if they want to switch their model from showing the effects of an influenza outbreak to a measles outbreak or any other disease with similar transmission dynamics. This is the case with Barrett et al. (2008)’s Episimdemics, Aleman et al. (2011)’s model for Ontario and Lee et al. (2008) model for Alleghany County, Pennsylvania.
Often a model will focus on a specific disease because there is some reason that a general model will not capture the disease spread accurately enough. This can be the case if the infection dynamics of a disease are not typical, for example Ebola can be transmitted at funerals and cholera is transmitted through drinking water. Agent-based models have been based on specific outbreaks such as the Ebola outbreak in Liberia. Not only does this model include specifics to how Ebola spreads, such as contact at funerals, but the model is specific to Liberia including the number of hospital beds that were used for Ebola patients over the course of the epidemic (Merler et al. 2015) However, many of the agent-based models that focus on a specific disease model influenza. The transmission dynamics of influenza are closer to a general model than some other diseases such as TB or malaria and as such a SIR type model can be used. These models either focus on a specific strain of influenza such as H1N1 or H5N1, or treat influenza generally (Friás-Martínez et al. 2011; Dibble et al. 2007; (Rakowski et al. 2010). Specific agent-based models can also be used to determine how given interventions affect the spread of a virus. Among other topics agent-based models have been created to determine the effects that the government mandates had on the spread of the H1N1 virus in Mexico and how vaccination programs affect the incidence rate of Human papillomavirus (HPV) in Denmark (Friás-Martínez et al. 2011; Olsen & Jepsen 2010).
Society Model
The next stage in the taxonomy is the society model. Society models can be described as specific if they were created to model and actual society using real data and general otherwise. It is most often the case that a specific disease model is paired with a specific society model, as the idea behind such a model is typically to capture the dynamics of a past or current outbreak. In order to do this it is not just necessary to accurately model the disease but the society as well. However, there are cases where a specific disease is paired with a general society model. This would occur in cases where the model might be used for disease dynamics research where an overly realistic society is not needed such as in Duan et al. (2013)’s model looking into the possibility of identifying super spreaders.
In some cases, such as with Barrett et al. (2008)’s EpiSimdemics a general disease model is integrated with a specific society model. Such a model would be used as a public health tool where the effects of any possible new outbreak can be modelled on the given society. These models are most often used for planning for future outbreaks or epidemics as a public health tool for decision making. EpiSimdemics is a model that was created to scale to social networks with 100 million individuals. The parameters of the model can be altered in order to model different infectious diseases. Another modelling tool was created for the Greater Toronto area in Canada to determine the best mitigation strategies in the case of a potential epidemic (Aleman et al. 2011).
General disease models can also be combined with a general society model. Although the results of the models cannot be directly applied to a given society, these types of models can be used for research purposes. For example, models can show how factors such as fleeing from fear might change the course of the outbreak and how the differences in the number of individuals in different categories (susceptible, infected and recovered) over time compared to the results of an equation-based model will broadly show the effect that spatial mixing has on the outbreak of a disease. The models by Epstein et al. (2008) and Dunham (2005) are both examples of these types of models.
Transportation Model
We consider two levels of transportation for the taxonomy: models with transport and models without transport. Although there could potentially be finer levels created based on the complexity in the transportation model we felt that the boundaries between these levels were too fuzzy to be useful.
Models without transport tend to be matched with specific disease models as not including transportation in the model is most useful when the disease dynamics can be modelled with contact network structures versus day to day interactions. This is often the case with blood/bodily fluid borne diseases such as HIV or HPV, for example Olsen & Jepsen (2010)’s model for the spread of HPV in Denmark. In such cases adding transportation into the model would only serve to slow the simulation down. Models without transport can be paired with either a specific or general society based on the aim of the model.
Models with transportation are paired with both general and specific disease models. The types of diseases that are transmitted with airborne transmission that can be substituted into a general model are often relatively contagious and a transportation model helps to better capture the spread of the disease by identifying random contacts outside of an agent’s family or friends. Epstein et al. (2008) use a general disease model and a transportation model to determine how agents fleeing during an epidemic will lead to greater spread of the disease. Similarly transportation can help capture the dynamics of disease spread for models of specific diseases. For example, Crooks & Hailegiorgis (2014) use agents’ movements to determine if, when and where an agent is drinking contaminated water that may lead to a cholera infection. Vector borne diseases can also be reliant on movement, if an agent travels to an area with a higher concentration of the vector population it will be more likely that the agent will be infected (Linard et al. 2009). Pairing a transport model with a specific disease model helps to capture dynamics of many infectious disease that may have been modelled incompletely without movement.
Similar to the models with no transport, models with transport can be matched with either specific or general society models. The choice of society would be determined by the researchers and their goals for the model. For example, Olsen & Jepsen (2010) wanted to study the effects of the HPV vaccine on the population of Denmark. To do this they needed to include a specific society because they wanted their results to be specific to Denmark, however, because the HPV is a sexually transmitted disease the model does not need to include transportation as daily interactions on the road or in work or school will not spread the disease.
Environmental Model
The taxonomy breaks the environmental model down to three levels: no added environment, maps, and maps plus other environmental factors. Other environmental factors could be anything from temperature and precipitation to a vector population or a hydrology model. Models with other environmental factors added into the model are usually combined with a specific disease model since the environmental factors added into the model should be factors that are related to the disease. For example, the model by Crooks & Hailegiorgis (2014) is a model for the spread of cholera in a specific refugee camp and includes hydrology models and precipitation models as they are essential to the spread of the disease. In most cases these models are also paired with a specific society since the environmental factors are based on what is seen in the real world. Models with environmental factors are also typically paired with a model with transportation. A simulation with specific disease, specific society and high level environment would create a simulation where the results can be easily applied to a real life scenario. However, the models will also be data heavy which could lead to slow initialization and computing time.
Environmental models that include roads, buildings and/or maps are nearly always paired with a specific society model. This makes sense in the idea that in order to add roads or a map in the environment the researchers would need to choose which map or which roads to use based on the society that is being modelled. Additionally, models with an environment that is made up of maps or roads will usually include transportation. There is not much point in creating an environment with roads if the agents do not move along them. Such models are matched with either a general or a specific disease model.
Models with no added environment are those where the simulation is solely made up of agents interacting with each other in an open space. These types of environments can be in simulations that do not include transportation but focus on a specific disease, such as Olsen & Jepsen (2010)’s model. Because the agents will not move through the environment there isn’t as much of a need to create any detail in the environment for the agents to interact with. Other models with no added environment will include transportation. Most often these models are dealing with a general society and either a specific or general disease model.
Applying the Taxonomy
Table 1 puts the 20 models analyzed in our literature review into the classification system. If the disease model or the society model is a specific model the name of the disease or society is also included in the table.
The table also includes the possible use of the model. Based on the models reviewed, there are four main uses for agent-based models of infectious diseases: disease dynamics research, agent-based modelling research, epidemic planning, and lessons learned. Disease dynamics research focuses on learning information about how a disease will transmit in a circumstance that would otherwise be hard to learn without a real life outbreak scenario. For example, how finding and treating super-spreaders can help to lessen an epidemic and how effective contact tracing can help stop outbreaks of TB (Duan et al. 2013; Tian et al. 2013). Agent-based modelling research is concerned with finding new ways to use agent-based models and new methods for creating agent-based models. For example, Bobashev et al. (2007) explore how to combine agent-based and equation based models. A model used for epidemic planning such as Barrett et al. (2008)’s Episimdemics model is created to learn the best strategies to deal with outbreaks prior to an outbreak occurring while lessons learned is the idea of modelling a past outbreak in order to learn from what happened in the past and to determine if the measures taken to stop the spread of the disease were successful. For example, Friás-Martínez et al. (2011) modelled the 2009 H1N1 outbreak in Mexico City in order to determine if restrictions on movement affected the course of the epidemic.
Using the table to find similarities in the models that have the same use can help to better use the taxonomy. For example, determining that a lessons learned model always contains a specific disease and specific society will direct researchers to that branch on the taxonomy and help them to make decisions on the other components they need to include in the model.
Models that focus on a specific society with either a general or specific disease tend to be used for epidemic planning. A number of the models reviewed, (Crooks & Hailegiorgis 2014; Rakowski et al. 2010), attempt to accurately simulate the spread of a specific disease so that the model could be used in the future to determine best practices. Other models such as, Barrett et al. (2008) and Aleman et al. (2011) use a general disease to create a model that can be used for multiple outbreaks. Results published from a study using the Simdemics model show that a combination of school closures, individual adaptive behaviour, and targeted antiviral distribution could reduce the impact of an influenza-like pandemic by 87% and the income loss from such a pandemic would decrease by 82% compared to a base case (Apolloni et al. 2009). The EpiSimdemics model is able to simulate detailed information on a disease spreading through a population including the individuals infected, where they were infected and who infected them. The information EpiSimdemics provides allows for identification of the severity of the epidemic as a whole and in certain subpopulations. The model has been used for multiple studies including those on pandemic planning for the US Department of Defence and the US Department of Health and Human Services. Looking at the effects of sequestering military sub-populations during a pandemic, the EpiSimdemics model determined that counter-intuitively sequestration may lead to more infections. It was determined this was because certain diseases can be infectious before being symptomatic and although overall contacts would decrease with sequestration contacts in a smaller group of individuals, those who were sharing military quarters, would increase: resulting in infectious individuals being in close contact with susceptible individuals for a long period of time (Barrett et al. 2008). The general disease model combined with specific society and transportation models allows for the user of the model to change the infection dynamics based on what situation they would like to study. This saves the effort of recreating a model for the same population every time a study needs to be done and gives the user the advantage of having a previously validated model. Another similar planning result obtained from a specific disease model is a cost-effectiveness analysis. Olsen & Jepsen (2010) use an agent-based model to determine cost-effectiveness ratios for HPV vaccinations and determine that while a new vaccination program will incur costs, in the long term it will save treatment costs and improve quality of life and survival.
| Paper | Disease | Society | Transport | Environment | Use |
| Ajelli et al. 2010 | General | Specific (Italy) | Yes | Maps | Agent-Based Model Research |
| Aleman et al. 2011 | General | Specific (Ontario) | Yes | Maps | Epidemic Planning |
| Barrett et al. 2008 | General | Specific (USA) | Yes | Maps | Epidemic Planning |
| Bobashev et al. 2007 | General | Specific (World) | Yes | No | Agent-Based Model Research |
| Crooks and Hailegiorgis 2014 | Specific (Cholera) | Specific (Dadaab refugee camp) | Yes | Maps plus | Epidemic Planning |
| Dibble et al. 2007 | Specific (H5N1) | Specific (USA) | Yes | None | Epidemic Planning |
| Duan et al. 2013 | Specific (SARS) | General | Yes | None | Disease Dynamics Research |
| Dunham 2005 | General | General | Yes | None | Disease Dynamics Research |
| Epstein et al. 2008 | General | General | Yes | None | Disease Dynamics Research |
| Frias-Martinez et al. 2011 | Specific (H1N1) | Specific (Mexico City) | Yes | Maps | Lessons Learned |
| Lee et al. 2010 | General | Specific (Allegheny, PA, USA) | Yes | Maps | Epidemic Planning |
| Linard et al. 2009 | Specific (Malaria) | Specific (South France) | Yes | Maps plus | Epidemic Planning |
| Mao 2014 | Specific (Influenza) | Specific (Bu alo, NY, USA) | Yes | Maps | Disease Dynamics Research |
| Merler et al. 2015 | Specific (Ebola) | Specific (Liberia) | Yes | Maps | Lessons Learned |
| Olsen and Jepsen 2010 | Specific (HPV) | Specific (Denmark) | No | None | Epidemic Planning |
| Perez and Dragicevic 2009 | General | Specific (Burnby, Canada) | Yes | Maps | Epidemic Planning |
| Rakowski et al. 2010 | Specific (Influenza) | Specific (Poland) | Yes | Maps | Epidemic Planning |
| Simoes 2006 | Specific (Mumps) | Specific (Portugal) | Yes | Maps | Lessons Learned |
| Skvortsov et al. 2007 | General | Specific (Australia town) | Yes | Maps | Epidemic Planning |
| Tian et al. 2013 | Specific (Tuberculosis) | Specific (Saskathewan, Canada) | No | None | Disease Dynamics Research |
If a researcher wished to create a model for epidemic planning they could go to the taxonomy and look at the branches that contain specific society. Looking at the taxonomy, if they also wished to include a general disease model they would know that a transportation model should be included and they would only need to decide on no added environment or maps. Alternatively if they wanted to include a specific disease model, the researcher could need to decide if they wanted to include transportation in their model based off of the transmission of the disease being modelled (human-to-human, food or water to human, vector to human). Deciding to not include transportation would also result in not including any added environment, while deciding to include transportation would require a decision on what level of environment would need to be added.
From the table, models that are for lessons learned from a past outbreak tend to be created with a specific disease and specific society model. For example, Frias-Martinez created a model for the H1N1 outbreak in Mexico city in order to evaluate the cities mitigation strategy (Friás-Martínez et al. 2011). Similarly, Merler et al. (2015) looked into the Ebola outbreak in Liberia to determine if safe funerals and Ebola patient only medical centers affected the outbreak. Knowing this, if a researcher wanted to create a model that would be used for lessons learned they could go to the taxonomy and follow the branch to specific disease and specific society. Based on the disease being modelled and the type of transmission, they could then decide if the model should include transportation and how much added environment to include.
Models that have a result focused on agent-based model research are often created to find a solution to some of the problems in the field of agent-based modelling for infectious disease epidemiology. One of the main barriers in the uptake of agent-based models is the time it can take to run a detailed simulation coupled with the large amount of processing power needed. In order to overcome this barrier experimentation must be done to create more efficient agent-based models. This is already occurring in cases such as Bobashev et al. (2007) where an agent-based model is combined with an equation based model to improve efficiency. As agent-based models become faster and more efficient, more detail will be able to be added to the models. Hopefully this will result in larger uptake of agent-based models to help determine policy and direct research. These types of models will usually have a general disease, which would then require the modellers to decide if they wanted a general or specific society and if they choose a specific society if they should add environment to the model.
Models that are created to look into disease dynamics research can be placed anywhere on the taxonomy. Results from disease dynamics research models include learning about the effects of super-spreaders on an outbreak (Duan et al. 2013), looking into the effects of fleeing an outbreak (Epstein et al. 2008), or what effect the spread of fear and knowledge of the outbreak will have Mao (2014). As these models can have a general or specific disease, a general or specific society, transportation or no transportation and any level of environment, to decide what will work best for their model a researcher should go through each layer of the taxonomy and determine what will work best for them. For example, to see the effects of fleeing on an outbreak, Epstein et al. (2008) decided on a general disease model as they were not focusing on a specific disease but wanted to look more generally at what happens and a general society again to see the general effects of fleeing that might occur in any society. Once a general disease and society were chosen the options on the taxonomy include transportation and no added environment.
Although we have fit all of the models we reviewed into our taxonomy we are aware that no taxonomy can be completely comprehensive and there may be models that do not fit nicely into our classification. Even if this occurs we feel that our taxonomy is still a useful tool as it will work for the majority of agent-based infectious disease epidemiology models and was created based on evidence from the literature. It should also be noted that the taxonomy is not all inclusive and that there are other characteristics of the simulations that could be included. The taxonomy should, however, aid readers and simulation designers alike in determining the use of a simulation based on the different components of the simulation are handled and what to expect from the results.
Conclusions
Agent-based models can be a useful tool in helping to stop or prevent the spread of an infectious disease. Models such as Barrett et al. (2008)’s Episimdemics are already being used to influence policy. Merler et al. (2015) and Friás-Martínez et al. (2011) have studied past outbreaks to determine the success of interventions to help inform in case of future outbreaks. However, in order to be effective a model should be based on appropriate data and should be validated - both of which can prove to be a challenge. Data accessibility is a major obstacle when creating an agent-based model. For example, although Friás-Martínez et al. (2011)’s use of cell phone location data made their transportation model extremely accurate the majority of researchers will not be able to access such a dataset easily. If agent-based models are to be routinely used as policy tools a consistent validation method should be determined. Without such a method it may be difficult to distinguish a model that will provide accurate results for a given population from a model that will not.
The freedom and flexibility in agent-based model design allows many different type of models to be created even just in the field of infectious disease epidemiology. Yet the lack of clear protocols in creating and describing agent-based models can lead to confusion in understanding the methodology of a given agent-based model. Because of this it is essential to understand the different types of agent-based epidemiology models and how they relate to each other. The literature shows that similarities among existing agent-based infectious disease epidemiology models exist and that there are different ways to compare the simulations. These comparisons tend to be driven by similarities or differences in the components of the model, disease, society transportation and environment, and how the model handles the components.
For both disease and society we found that models in the literature tend to create either specific components based on data or general components where parameters can be adjusted to model multiple diseases or results can be applied to any society. The choice of general or specific disease model or a general or specific society model will have an effect on the transportation and environment components used, advantages and disadvantages of the model, possible uses of the model and the validation process.
As there are many possible combinations of the disease, society, transportation, and environment components of a model, each with potentially different uses, validation techniques, advantages and disadvantages we felt that the current literature was missing a classification tool. Using the knowledge we gained from our literature review we formulated our taxonomy. The taxonomy should aid readers and modellers alike in determining the use of a model based on how the different components of the simulation fit together. One of the problems with the current agent-based modelling literature is the lack of clear definitions and standards for agent-based models in infectious disease epidemiology and the components of those models due to the flexibility and freedom allowed in model design. We feel that creating a taxonomy can help to classify agent-based infectious disease epidemiological models and is a move towards solving the problem of definition without sacrificing the flexibility that attracts researchers to the agent-based modelling field.
In addition to helping classify the existing models in the literature we feel that the taxonomy can help researchers in creating models through determining which components are necessary for their intended use. For example, if a model is being created for epidemic planning it will need to have a specific society component. Once the components of a model are determined the taxonomy can help researchers identify the available methods for validation. For instance, if a general disease model or general society is used it may not be possible to compare the results to past outbreaks, while the use of a specific disease and a specific society makes the use of past outbreaks as a validation method a possibility. We see this as a real benefit of the taxonomy. By helping researchers identify the range of validation techniques that are suitable for a specific model the taxonomy can help standardise the approaches to validation that are used for agent based models for epidemiology. As discussed earlier in this article, the fact that different researchers employ very different levels of validation of their models is a recognised issue in agent-based modelling research so something to help standardisation could be a real benefit.
One of the biggest restrictions in agent-based modelling is computing power. Availability of computing resources can have a large role in determining the level of detail of a model. In order to create more complex and realistic models this limitation will need to be overcome. With recent advances in computing power, this is already becoming a reality and the level of complexity in agent-based modelling is far surpassing that of past decades. As the use of techniques taking advantage of these advances increase the field of agent-based modelling should become more rich. For example, cloud computing allows access to high performance computing clusters and when utilized by agent-based modellers will provide more accurate faster results (Taylor et al. 2014). This may allow modellers to no longer have to simplify different aspects of their model and will give them more freedom and flexibility in model creation making a taxonomy for model classification all the more important.
Acknowledgements
This work was partly supported by the Fíosraigh Scholarship Programme of the Dublin Institute of Technology.References
AJELLI, M., Goncalves, B., Balcan, D., Colizza, V., Hu, H., Ramasco, J. J., Merler, S. & Vespignani, A. (2010). Comparing large-scale computational approaches to epidemic modeling: Agent-based versus structured metapopulation models. BMC Infectious Diseases, 10(190). [doi:10.1186/1471-2334-10-190 ]
ALEMAN, D. M., Wibisono, T. G. & Schwartz, B. (2011). A nonomogeneous agent-based simulation approach to modeling the spread of disease in a pandemic. Informs, 41(3), 301–315.
APOLLONI, A., Kumar, V. A., Marathe, M. V. & Swarup, S. (2009). Computational epidemiology in a connected world. IEEE Computer Society, (pp. 97–100). [doi:10.1109/MC.2009.386 ]
BARRETT, C. L., Bisset, K. R., Eubank, S. G., Feng, X. & Marathe, M. V. (2008). Episimdemics: An efficient algorithm for simulating the spread of infectious disease over large realistic social networks. International Conference for High Performance Computing, Networking, Storage and Analysis.
BOBASHEV, G. V., Goedecke, D. M., Yu, F. & Epstein, J. M. (2007). A hybrid epidemic model: Combining the advantages of agent-based and equation based-approaches. Proceedings of the 2007 Winter Simulation Conference, (pp. 1532–1537). [doi:10.1109/WSC.2007.4419767 ]
BRUCH, E. & Atwell, J. (2015). Agent-based models in empirical social research. Sociological Methods & Research, 44(2), 186–221.
CROOKS, A. T. & Hailegiorgis, A. B. (2014). An agent-based modeling approach applied to the spread of cholera. Environmental Modelling & Software, 62, 164 – 177. [doi:10.1016/j.envsoft.2014.08.027 ]
DIBBLE, C., Wendel, S. & Carle, K. (2007). Simulating pandemic influenza risks of US cities. Proceedings of the 2007 Winter Simulation Conference, (pp. 1548–1550).
DUAN, W., Qiu, X., Cao, Z., Zheng, X. & Cui, K. (2013). Heterogeneous and stochastic agent-based models for analyzing infectious diseases’ super spreaders. IEEE Intelligent Systems, 13, 1541–1672. [doi:10.1109/mis.2013.29 ]
DUNHAM, J. B. (2005). An agent-based spatially explicit epidemiological model in MASON. Journal of Artificial Societies and Social Simulation, 9(1), 3: https://www.jasss.org/9/1/3.html.
EPSTEIN, J. M., Parker, J., Cummings, D. & Hammond, R. A. (2008). Coupled contagion dynamics of fear and disease: Mathematical and computational explorations. PLoS ONE, 3(12), 1–11. [doi:10.1371/journal.pone.0003955 ]
FRIÁS-MARTÍNEZ, E., Williamson, G. & Friás-Martínez, V. (2011). An agent-based model of epidemic spread using human mobility and social network information. IEEE Conference on Social Computing.
HERNÁN, M. A. (2014). Invited commentary: Agent-based models for casual inference–reweighting data and theory in epidemiology. American Journal of Epidemiology, 181(2), 103–105. [doi:10.1093/aje/kwu272 ]
HOMER, J. B. & Hirsch, G. B. (2006). System dynamics modeling for public health: Background and opportunities. American Journal of Public Health, 96(3), 452–458.
KEELING, M. J. & Rohani, P. (2008). Modeling Infectious Diseases in Humans and Animals. Princeton, NJ: Princeton University Press.
LEE, B. Y., Brown, S. T., Cooley, P., Potter, M. A., Wheaton, W. D., Voorhees, R. E., Stebbins, S., Grefenstette, J. J., Zimmer, S. M., Zimmerman, R., Assi, T.-M., Bailey, R. R., Wagener, D. K. & Burke, D. S. (2008). Simulating school closure strategies to mitigate an influenza epidemic. Journal of Public Health Management and Practice, 16(3), 252–261.
LINARD, C., Poncon, N., Fontenille, D. & Lambin, E. F. (2009). A multi-agent simulation to assess the risk of malaria re-emergence in Southern France. Ecological Modelling, 220(2), 160 – 174. [doi:10.1016/j.ecolmodel.2008.09.001 ]
MAC NAMEE, B. & Cunningham, P. (2003). Creating socially interactive no-player characters: The µ-SIV system. International Journal of Intelligent Games and Simulation, 2(1), 186–221: http://www.comp.dit.ie/bmacnamee/papers/uSIC.pdf.
MAO, L. (2014). Modeling triple-diffusions of infectious diseases, information, and preventive behaviors through a metropolitan social network – an agent-based simulation. Applied Geography, 50, 31 – 39. [doi:10.1016/j.apgeog.2014.02.005 ]
MERLER, S., Ajelli, M., Fumanelli, L., Gomes, M. F. C., y Piontti, A. P., Rossi, L., Chao, D. L., Jr, I. M. L., Halloran, M. E. & Vespignani, A. (2015). Spatiotemporal spread of the 2014 outbreak of Ebola virus disease in Liberia and the effectiveness of non-pharmaceutical interventions: A computational modelling analysis. The Lancet Infectious Diseases, 15(2), 204 – 211.
OLSEN, J. & Jepsen, M. R. (2010). Human papillomavirus transmission and cost-effectiveness of introducing quadrivalent HPV vaccination in Denmark. International Journal of Technology Assesment in Health Care, 26(2), 183 – 191. [doi:10.1017/S0266462310000085 ]
PEREZ, L. & Dragicevic, S. (2009). An agent-based approach for modeling dynamics of contagious disease spread. International Journal of Health Geographics, 8(50), 1–17.
RAKOWSKI, F., Gruziel, M., Bieniasz Krzywiec, L. & Radomski, J. P. (2010). Influenza epidemic spread simulation for Poland at a large scale, individual based model study. Physica A: Statistical Mechanics and its Applications, 389(16), 3149-3165. [doi:10.1016/j.physa.2010.04.029 ]
RICHIARDI, M., Leombruni, R., Saam, N. J. & Sonnessa, M. (2006). A common protocol for agent-based social simulation. Journal of Artificial Societies and Social Simulation, 9(1), 15: https://www.jasss.org/9/1/15.html.
SIMOES, J. M. (2006). Modelling a mumps outbreak through spatially explicit agents. Potentials of Complexity Science for Business, Governments, and the Media 2006: http://www.casa.ucl.ac.uk/joanamargarida/papers/SimoesJ.pdf.
SKVORTSOV, A. T., Connell, R. B., Dawson, P. D. & Gailis, R. M. (2007). Epidemic modelling: Validation of agent-based simulation by using simple mathematical models. International Congress on Simulation and Modelling, (pp. 657–662): http://mssanz.org.au.previewdns.com/MODSIM07/papers/13_s20/EpidemicModeling_s20_Skvortsov_.pdf.
TAYLOR, S. J. E., Kiss, T. & Anagnostou, A. (2014). A tutorial on cloud computing for agent-based modeling and simulation with Repast. Proceedings of the 2014 Winter Simulation Conference. [doi:10.1109/WSC.2014.7019888 ]
TIAN, Y., Osgood, N. D., Al Azem, A. & Hoeppner, V. H. (2013). Evaluating the effectiveness of contact tracing on tuberculosis outcomes in Saskatchean using individual-based modeling. Health Education and Behavior, 40(15), 985–1105.
XIA, H., Barrett, C., Chen, J. & Marathe, M. V. (2013). Computational methods for testing adequacy and quality of massive synthetic proximity social networks. CSE ’13 Proceedings of the 2013 IEEE 16th International Conference on Computational Science and Engineering, (pp. 1113–1120): http://staff.vbi.vt.edu/chenj/pub/NDSSL-TR-13-153.pdf.
