Kerry L. Shaw and Kyle Wagner (2008)
Cricketsim: a Genetic and Evolutionary Computer Simulation
Journal of Artificial Societies and Social Simulation
vol. 11, no. 1 3
<https://www.jasss.org/11/1/3.html>
For information about citing this article, click here
Received: 20-Apr-2007 Accepted: 25-Oct-2007 Published: 31-Jan-2008
 Abstract
Abstract
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Figure 1. A short example genome from a male hybrid organism. Chromosomal location refers to the location on the chromosome of 100.0 units. The sex locus is positioned at 99.0 units. The signal loci are positioned at 0.5 and 57.2 units. The preference loci are positioned at 65.9 and 74.1 units. The fitness loci (for internal and external lifespan penalties) share the remaining loci at 87.0, 87.2, and 87.4 units. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

|
| Figure 2. Screenshot of a 40×40 world after 25 timesteps, showing male and female densities in each cell (males are red, females are green, cells with a mixture have an interpolated color value between red and green). Black areas indicate that no crickets are in that cell. This is the "micro" mode display, showing information only about male/female densities. Each population began in a tightly-clustered area and has started to grow outwards. |
|
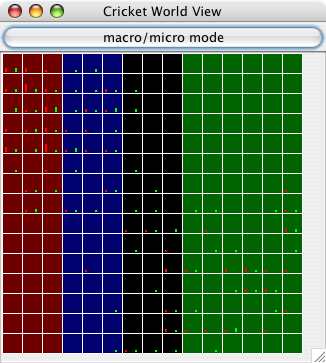
| Figure 3. Screenshot of a 15×15 world after a few timesteps have elapsed. This is the "macro" mode display, which shows more information about each cell. Each cell displays its terrain type as a color (there are four terrain types in this particular run). Also, each cell has a bar whose height corresponds to the number of males (red) and females (green) in that cell. Notice that the fringes of each population cluster tend to be male-only or female-only and sparsely populated, while the center of each population (upper left and lower right) contains many males and females. |
|
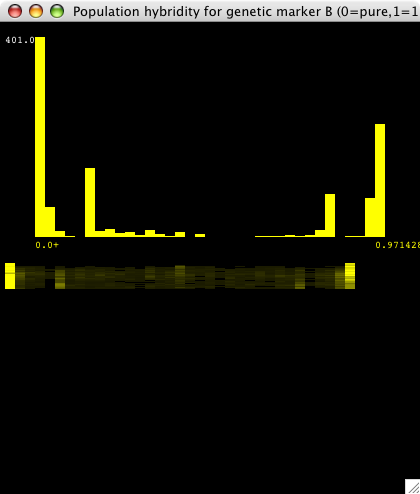
| Figure 4. This is a real-time histogram of hybridity with respect to the "B" genetic marker (the marker for population 2). The tall bars at either end indicate that most individuals are either pure B (at 0.0) or pure A (at 0.97, which is a histogram bin containing 0.97 to 1.0 hybridity individuals). There are some significant hybrid populations as well. The "smear graph" below shows slices of the above histogram over time, with t=0 at the top. At t=0, and for a while, there are no yellow bars in between 0.0 and 1.0 except at the ends, so both populations are pure. As time goes on, some hybrid matings occur, which is reflected by the dim yellow bars that appear. The intensity of the bars corresponds to the height of the histogram at that point. This smear graph shows how the hybridity histogram changes over time. |
|
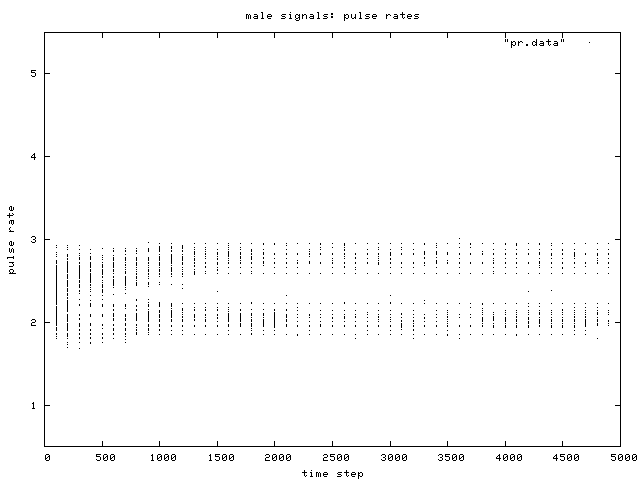
| Figure 5. Scatterplot of individual male signal pulse rate values over time, for one simulation run. Two phenotypic groups are clear after about generation 500. |
|
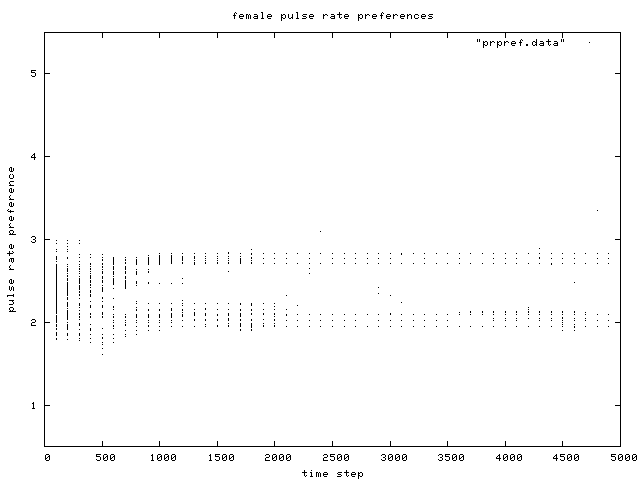
| Figure 6. Scatterplot of individual female pulse rate (signal) preferences over time, for the same simulation run as in Figure 1. Like male pulse rate, two preference groups are clear after about generation 500. |
|
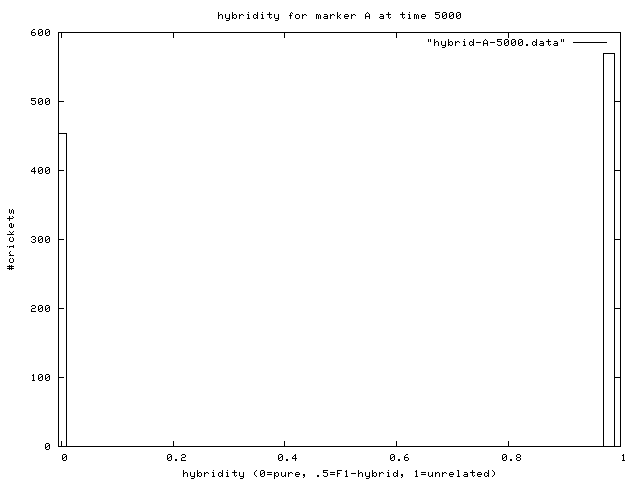
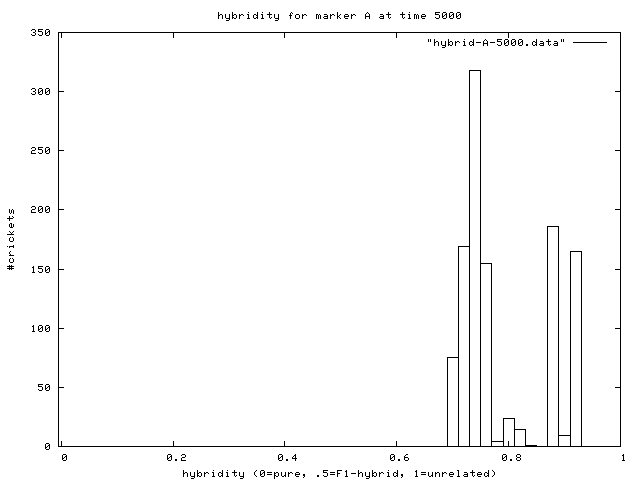
| Figure 7. Hybridity of one experimental population at end of simulation. Pure A (right stack) and B (left stack) individuals are evident. |
| ||||||
| Figure 8. Initial and final spatial distribution of population types (A=more than 50% genes from A population, B=more than 50% genes from B population). | ||||||
|
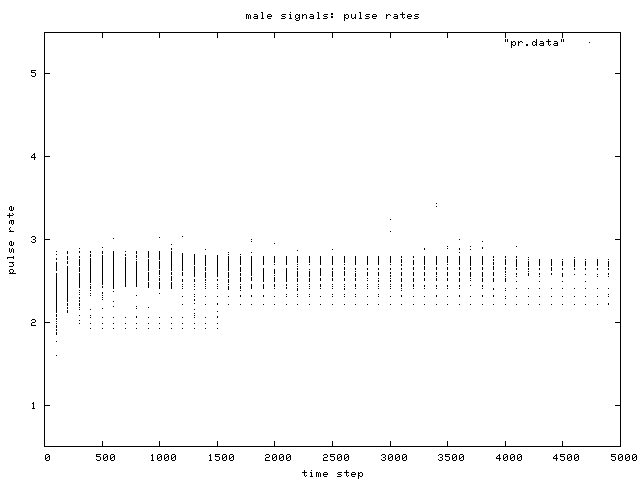
| Figure 9. Scatterplot of individual male signal pulse rates over time, for one simulation run, control condition. |
|
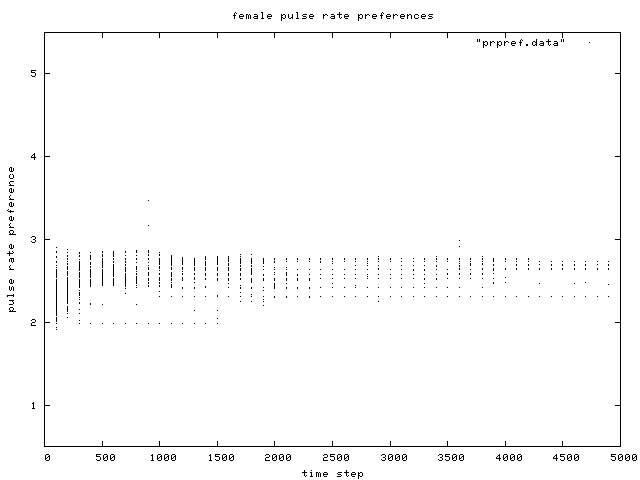
| Figure 10. Scatterplot of individual female pulse rate (signal) preferences over time, for the same control condition simulation run as in Figure 6. |
|
| Figure 11. Hybridity of one control population at end of simulation. The hybridity of individuals is distinctly bimodal, although no splitting was evident in the signal and preference phenotypes. |
2 Big-O notation expresses the order of magnitude of work that an algorithm must perform. O(N) would mean that an algorithm operating on N items would require cN steps, where c is a numerical constant. Given small enough values of c (quite typically the case), an O(N) algorithm is much better than an O(N^2) algorithm.
3 Please refer to http://www.bluegradient.org/cricketsim/cricketsim.htmlfor a full description of these parameters as well as the many other parameters which can be set in the program.
BRIDEAU, N J, Flores, H A, Wang, J, Maheshwari, S, Wang, X, Barbash, D A (2006). 'Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila'. Nature 314: 1292-1295.
DEANGELIS, D L, Mooij, W M (2005). 'Individual-based modeling of ecological and evolutionary processes'. Annual Reviews of Ecology and Systematics 36: 147-168.
DE BOURCIER, P (1996) 'Using a-life to study bee life: the economics of central place foraging'. In Maes, P et al. From Animals to Animats 4 ed. Bradford/MIT, MA.
DOEBILI, M, Deickmann, U (2000). 'Evolutionary branching and sympatric speciation caused by different types of ecological interactions'. American Naturalist 156: S77-S101.
GHEORGHE, M, Holcombe, M, Kefalas, P (2001). 'Computational models of collective foraging'. BioSystems 61: 133-141.
GRIMM, V, Railsback, S F (2005) Individual based modeling and ecology. Princeton, NJ: Princeton University Press.
HENSON, S M, Costantino, R F, Cushing, J M, Desharnais, R A, Dennis, B, King, A A (2001). 'Lattice effects observed in chaotic dynamics of experimental populations'. Science 294: 602-605.
HILSCHER, R (2005). 'Agent-based models of competitive speciation I: effects of mate search tactics and ecological conditions'. Evolutionary Ecology Research 7: 943-971.
IKEGAMI, T, Kaneko, K (1990). 'Computer symbiosis: emergence of symbiotic behavior through evolution'. Physica D 42: 235-243.
LOTKA, A J (1925) Elements of physical biology. Baltimore, MD: Williams & Wilkins Publishing.
MEAD, L S, Arnold, S J (2004). 'Quantitative genetic models of sexual selection'. Trends in Ecology and Evolution 19: 264-271.
MENDELSON, T C, Shaw, K L (2005). 'Rapid speciation in an arthropod'. Nature 433: 375-376.
OTTE, D (1994) The Crickets of Hawaii: Origin, Systematics, and Evolution. Orthoptera Society/Academy of Natural Sciences of Philadelphia.
SCHMITZ, O J (2000). 'Combining field experiments and individual-based modeling to identify the dynamically relevant organizational scale in a field system'. Oikos 89: 471-484.
SADEDIN, S, Littlejohn, M J (2003). 'A spatially explicit individual-based model of reinforcement in hybrid zones'. Evolution 57: 962-970.
SERVEDIO, M R (2000). 'Reinforcement and the genetics of nonrandom mating'. Evolution 54: 21-29.
SEYFARTH, R M (1977). 'A model of social grooming among adult female monkeys'. Journal of Theoretical Biology 65: 671-698.
SHAW, K L (2000). 'Further acoustic diversity in Hawaiian forests: Two new species of Hawaiian cricket (Orthoptera: Gryllidae: Laupala)'. Zool. J. Linn. Soc. 129: 73-91.
SHAW, K L, Herlihy, D P (2000). 'Acoustic preference functions and song variability in the Hawaiian cricket Laupala cerasina'. Proceedings of the Royal Society of London B 267: 577-584.
STRAND, E, Huse, G, Giske, J (2002). 'Artificial evolution of life history and behavior'. American Naturalist 159: 624-644.
VENTRELLA, J (1996) "Sexual swimmers: emergent morphology and locomotion without a fitness function". In Maes, P, Mataric M J, Meyer J-A, Pollack J, Wilson, S W (Eds.) From Animals to Animats 4, Bradford/MIT, MA. Pp 484-496.
VOLTERRA, V (1926). 'Variazioni e fluttuazioni del numero d'individui in specie animali conviventi'. Mem. R. Accad. Naz. dei Lincei. Ser. VI(2).
WAGNER, K, Reggia, J (2002). 'Evolving consensus among a population of communicators'. Complexity International, 9, http://www.complexity.org.au/ci/vol09/wagner01/wagner01.html.
WAGNER, K, Reggia, J, Wilkinson, G, Uriagereka, J (2001). 'Conditions enabling the emergence of inter-agent signaling in an artificial world'. Artificial Life 7: 3-32.
WAY, E (2001). 'The role of computation in modeling evolution'. BioSystems 60: 85-94.
Return to Contents of this issue
© Copyright Journal of Artificial Societies and Social Simulation, [2008]